Nest (protein structural motif)
The Nest is a type of protein structural motif. It is a small recurring anion-binding feature of both proteins and peptides.[1][2][3][4][5][6][7][8][9] Each consists of the main chain atoms of three consecutive amino acid residues. The main chain NH groups bind the anions while the side chain atoms are often not involved. Proline residues lack NH groups so are rare in nests. About one in 12 of amino acid residues in proteins, on average, belongs to a nest.
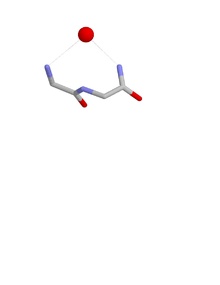
Nest conformations
[edit]The conformation of a nest is such that the NH groups of the first and third amino acid residues are liable to be hydrogen bonded to a negatively charged, or partially negatively charged, atom, often an oxygen atom. The NH of the second residue may also be hydrogen bonded to the same atom but usually points somewhat away. These main chain atoms form a concavity called a nest into which an anionic atom fits. Such anionic atoms are sometimes called eggs and more than one egg may occur bound to a nest. The oxyanion hole of the intestinal serine proteases is a functional example of a nest. Another occurs at the bottom of a deep cavity in the antibiotic peptide vancomycin which binds a key carboxylate group utilized during the final stages of bacterial cell wall synthesis, thereby preventing bacterial cells from multiplying.
Nests are defined by the conformation of the main chain atoms, namely the phi, psi dihedral angles of the first two amino acids in the nest. For a typical (RL) nest phii=-90°; psii=0°; phii+1=80°; psii+1=20°.
Nests vary in their degree of concavity. A few have so little that the concavity is lost; these peptides often bind cations via their main chain CO groups, instead of anions via their NH groups. The specificity filter of the potassium channel[10] and the water channel of aquaporin exhibit this more linear conformation in which carbonyl groups are employed by proteins to transport molecules across membranes. This near-linear conformation is also that found in a strand of alpha sheet[11][12][13]
Compound nests
[edit]If two nests overlap such that residue i+1 of the first nest is residue i of the second nest, a compound nest is formed. This has four NH groups instead of three. If three nests overlap such that residues i+1 and i+2 of the first nest are residue i of the second and third nest, a wider compound nest is formed with five NH groups, and so on. The main chain atoms form part of an incomplete ring with the NH groups all pointing roughly towards the centre of the ring. Because their concavities are often wider than simple nests, compound nests are commonly employed by proteins for binding multi-atom anions such as phosphates, as in the P-loop or Walker motifs, and in iron-sulphur clusters. The synthesized peptide Ser-Gly-Ala-Gly-Lys-Thr, designed as a minimal peptide P-loop, was shown to bind inorganic phosphate strongly at neutral pH.[14]
Types of nest
[edit]Simple nests are of two kinds called RL and LR depending on the sign of the phi angles of the first two nest residues. R residues have negative phi values (as in right-handed alpha-helices) and L residues have positive phi values (as in the left-handed alpha helix). Eighty percent of nests are RL and 20% are LR. When two nests overlap they may be RLR or LRL. When three nests overlap they may be RLRL or LRLR, and so on.
Every Schellman loop incorporates an RL nest in the last three of its six residues. The nest binds carbonyl oxygen atoms preceding it in sequence.
A number of antibody proteins have RLR nests within the hairpin loops of their H-chain CDRs (complementarity determining regions) bound to a carboxylate side chain. These have been engineered to give rise to monoclonal nest-containing antibodies specific for proteins with phosphorylated serines and threonines.[15]
Most PDZ domains have an RL nest at the beginning of the first beta-strand, with the function of recognizing the carboxylate group at the C-terminus of the domain's peptide or protein ligand.[16]
References
[edit]- ^ Watson, JD; Milner-White (2002). "A novel main-chain anion-binding site in proteins: The nest. A particular combination of phi,psi values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions". Journal of Molecular Biology. 315 (2): 171–182. doi:10.1006/jmbi.2001.5227. PMID 11779237.
- ^ Pal, D; Suhnel (2002). "New principles of protein structure: nests, eggs and what next?". Angew Chem Int Ed. 41 (24): 4663–4665. doi:10.1002/anie.200290009. PMID 12481319.
- ^ Milner-White, EJ; Nissink (2004). "Recurring main-chain anion-binding motifs in short polypeptides: nests". Acta Crystallographica Section D. D60 (11): 1935–1942. doi:10.1107/s0907444904021390. PMID 15502299.
- ^ Pajewski, R; Ferdani (2005). "Cation Dependence of Chloride Ion Complexation by Open-Chained Receptor Molecules in Chloroform Solution". Journal of the American Chemical Society. 127 (51): 18281–18295. doi:10.1021/ja0558894. PMID 16366583.
- ^ Berkessel, A; Koch (2006). "Asymmetric enone epoxidation by solid-phase bound peptides: further evidence for catalyst helicity and catalytic activity of individual strands". Biopolymers. 84 (1): 90–96. doi:10.1002/bip.20413. PMID 16283656.
- ^ Milner-White, EJ; Russell (2006). "Predicting the conformations of proteins and peptides in early evolution". Biology Direct. 3: 3. doi:10.1186/1745-6150-3-3. PMC 2241844. PMID 18226248.
- ^ Watson, JD; Laskowski (2005). "ProFunc: a server for predicting protein function from 3D structure". Nucleic Acids Research. 33 (Web Server): W89–W93. doi:10.1093/nar/gki414. PMC 1160175. PMID 15980588.
- ^ Langton, MJ; Serpell CJ; Beer PD (2016). "Anion recognition in water: Recent advances from a supramolecular and macromolecular perspective". Angewandte Chemie International Edition. 55 (6): 1974–1987. doi:10.1002/anie.201506589. PMC 4755225. PMID 26612067.
- ^ Cremer, P; Flood AS; Gibb BC; Mobley DL (2018). "Collaborative routes to clarifying the murky waters of aquaeous supramolcular chemistry". Nature Chemistry. 10 (1): 8–16. doi:10.1038/nchem.2894. PMID 29256514. S2CID 205298633.
- ^ Watson, JD; Milner-White (2002). "The conformations of polypeptide chains where the main-chain parts of successive residues are enantiomeric. Their occurrence in cation and anion-binding regions of proteins". Journal of Molecular Biology. 315 (15): 183–191. doi:10.1006/jmbi.2001.5228. PMID 11779238.
- ^ Milner-White, EJ; Watson (2006). "Amyloid formation may involve alpha- to beta sheet interconversion via peptide plane flipping". Structure. 14 (9): 1369–1376. doi:10.1016/j.str.2006.06.016. PMID 16962968.
- ^ Hayward, S; Milner-White (2008). "The geometry of α-sheet: Implications for its possible function as amyloid precursor in proteins". Proteins. 71 (1): 415–425. doi:10.1002/prot.21717. PMID 17957773. S2CID 43848293.
- ^ Hayward, S; Milner-White (2011). "Simulation of the β- to α-sheet transition results in a twisted sheet for antiparallel and an α-nanotube for parallel strands: implications for amyloid formation". Proteins. 79 (11): 3193–3207. doi:10.1002/prot.23154. PMID 21989939. S2CID 8761012.
- ^ Bianchi, A; Giorgi A; Ruzza P; Toniolo C (2013). "A synthetic hexapeptide designed to resemble a proteinaceous P-loop nest is shown to bind inorganic phosphate". Proteins. 80 (5): 1418–1424. doi:10.1002/prot.24038. PMID 22275093. S2CID 5401588.
- ^ Koerber, JT; Thomsen ND; Hannigan BT; DeGrado WF; Wells JA (2013). "Nature-inspired design of motif-specific antibody scaffolds". Nature Biotechnology. 31 (10): 916–921. doi:10.1038/nbt.2672. PMC 3795957. PMID 23955275.
- ^ Lee, H-J; Zheng JJ (2010). "PDZ Domains and their binding Partners: Structure Specificity and Modification". Cell Communication and Signaling. 8: 8. doi:10.1186/1478-811x-8-8. PMC 2891790. PMID 20509869.
External links
[edit]- ^ Leader, DP; Milner-White (2009). "Motivated Proteins: A web application for studying small three-dimensional protein motifs". BMC Bioinformatics. 10 (1): 60. doi:10.1186/1471-2105-10-60. PMC 2651126. PMID 19210785.
- ^ Golovin, A; Henrick (2008). "MSDmotif: exploring protein sites and motifs". BMC Bioinformatics. 9 (1): 312. doi:10.1186/1471-2105-9-312. PMC 2491636. PMID 18637174.
